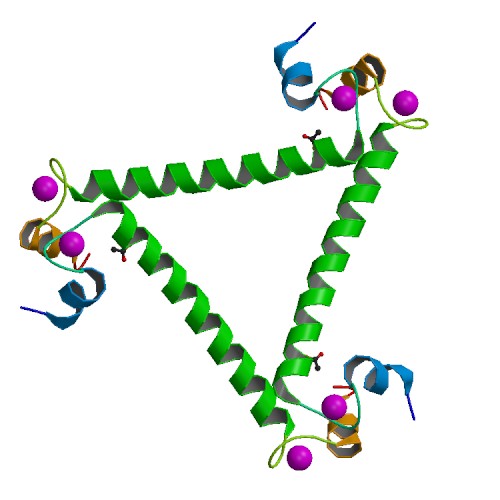
 |
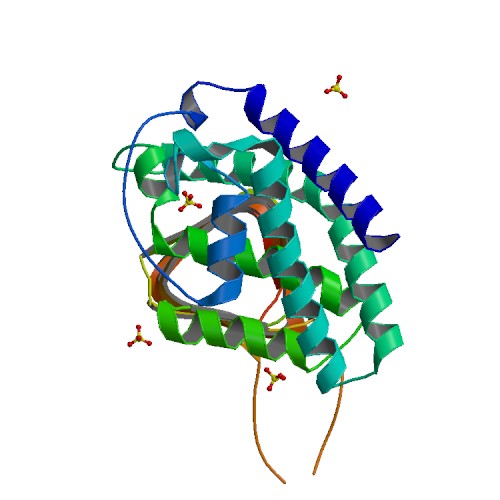
Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Lead
DOI: 10.2210/pdb3QJK/pdb
Classification: METAL BINDING PROTEIN
Organism(s): Entamoeba histolytica
Expression System: Escherichia coli
Deposited: 2011-01-29 Released: 2012-04-11
Deposition Author(s): S. Kumar, S. Gourinath
|
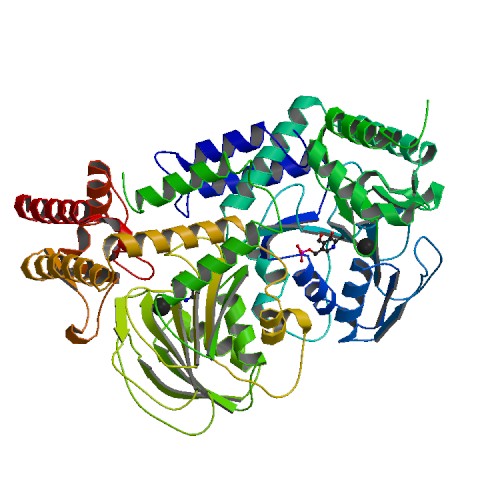 |
Crystal Structure of Delta 4 mutant of EhPSAT (Phosphoserine aminotransferase of Entamoeba histolytica)
DOI: 10.2210/pdb5YD2/pdb
Classification: TRANSFERASE
Organism(s): Entamoeba histolytica HM-3:IMSS
Expression System: Escherichia coli
Deposited: 2017-09-10 Released: 2018-10-31
Deposition Author(s): Singh, R.K., Gourinath, S.
|
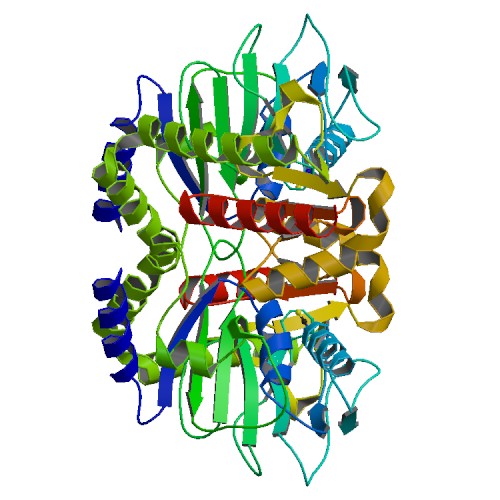 |
Crystal Structure of 45 amino acid deleted from N-terminal of Phosphoserine Aminotransferase (PSAT) of Entamoeba histolytica
DOI: 10.2210/pdb5YII/pdb
Classification: TRANSFERASE
Organism(s): Entamoeba histolytica
Expression System: Escherichia coli
Deposited: 2017-10-04 Released: 2018-10-24
Deposition Author(s): Singh, R.K., Gourinath, S.
|
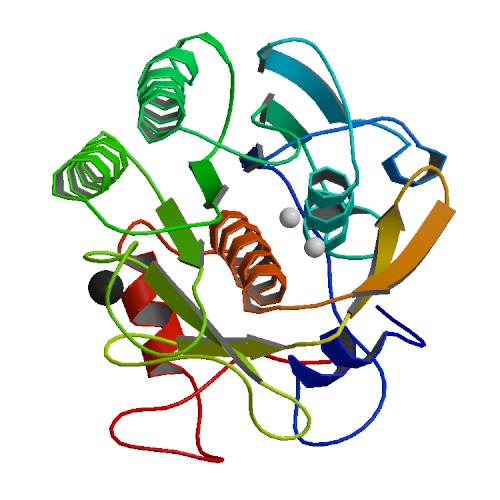 |
Mercury Induced Modifications in the Stereochemistry of the Active Site through Cys-73 in a Serine protease: Crystal Structure of the complex of a partially modified Proteinase K with Mercury at 1.8 A Resolution
DOI: 10.2210/pdb1HT3/pdb
Classification: HYDROLASE
Organism(s): Parengyodontium album
Deposited: 2000-12-27 Released: 2001-06-27
Deposition Author(s): Gourinath, S.
|
 |
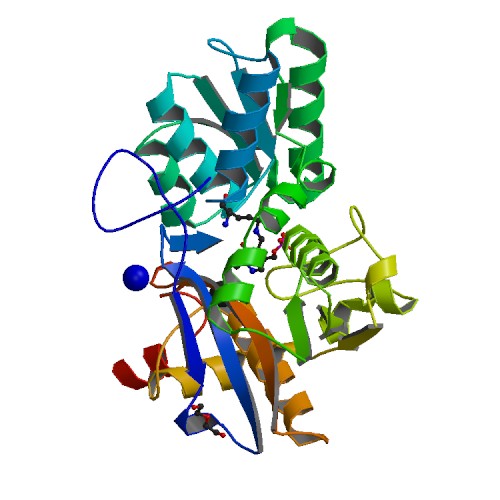
Crystal structure of the native serine acetyltransferase 1 from Entamoeba histolytica
DOI: 10.2210/pdb3P1B/pdb
Classification: TRANSFERASE
Organism(s): Entamoeba histolytica
Expression System: Escherichia coli
Deposited: 2010-09-30 Released: 2011-02-02
Deposition Author(s): Kumar, S., Gourinath, S.
|
 |
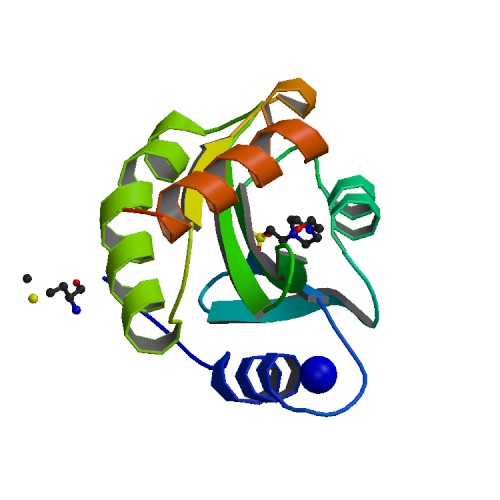
Crystal structure of O-Acetyl Serine Sulfhydrylase from Leishmania donovani
DOI: 10.2210/pdb3SPX/pdb
Classification: TRANSFERASE
Organism(s): Leishmania donovani
Expression System: Escherichia coli
Deposited: 2011-07-04 Released: 2012-07-04
Deposition Author(s): Raj, I., Gourinath, S.
|
 |
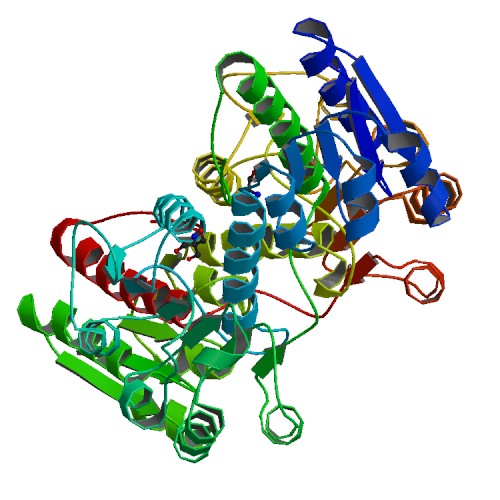
Crystal structure of coactosin from Entamoeba histolytica
DOI: 10.2210/pdb4LIZ/pdb
Classification: STRUCTURAL PROTEIN
Organism(s): Entamoeba histolytica HM-1:IMSS-A
Expression System: Escherichia coli
Deposited: 2013-07-04 Released: 2014-07-23
Deposition Author(s): Gourinath, S., Kumar, N.
|
 |
Crystal Structure of 3-phosphoglycerate Dehydrogenase from Entamoeba histolytica
DOI: 10.2210/pdb4NFY/pdb
Classification: OXIDOREDUCTASE
Organism(s): Entamoeba histolytica
Expression System: Escherichia coli
Deposited: 2013-11-01 Released: 2014-10-08
Deposition Author(s): Singh, R.K., Gourinath, S.
|
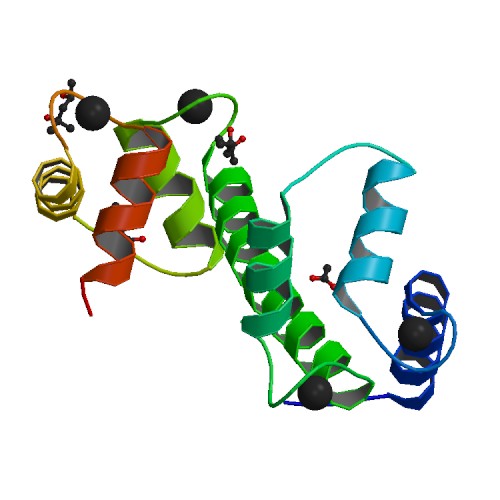 |
Crystal Structure of Arabidopsis thaliana Calmodulin-7
DOI: 10.2210/pdb5A2H/pdb
Classification: CALCIUM-BINDING PROTEIN
Organism(s): Arabidopsis thaliana
Expression System: Escherichia coli BL21(DE3)
Deposited: 2015-05-19 Released: 2016-03-30
Deposition Author(s): Kumar, S., Gourinath, S.
|
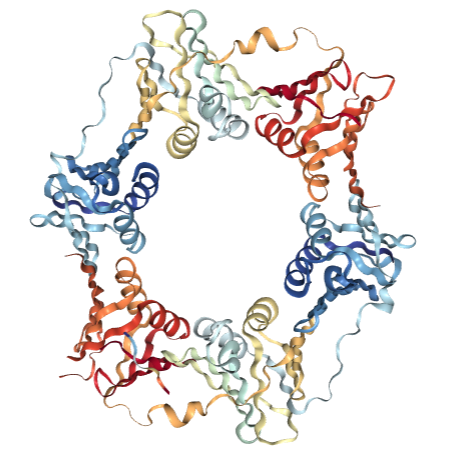 |
Crystal Structure of Helicobacter pylori beta clamp bound to DNA ligase peptide
DOI: 10.2210/pdb5FRQ/pdb
Classification: TRANSFERASE
Organism(s): Helicobacter pylori
Expression System: Escherichia coli BL21
Deposited: 2015-12-21 Released: 2016-08-17
Deposition Author(s): Pandey, P., Gourinath, S.
|
 |
The Crystal Structure of O-acetyl serine sulfhydralase from Brucella abortus
DOI: 10.2210/pdb5JIS/pdb
Classification: TRANSFERASE
Organism(s): Brucella abortus (strain S19)
Expression System: Escherichia coli
Deposited: 2016-04-22 Released: 2017-04-05
Deposition Author(s): Dharavath, S., Gourinath, S.
|
 |
Crystal Structure of PEG-bound SH3 domain of Myosin IB from Entamoeba histolytica
DOI: 10.2210/pdb5XG9/pdb
Classification: CONTRACTILE PROTEIN
Organism(s): Entamoeba histolytica
Expression System: Escherichia coli BL21(DE3)
Deposited: 2017-04-13 Released: 2017-08-16
Deposition Author(s): Gautam, G., Gourinath, S.
Funding Organization(s): SERB
|
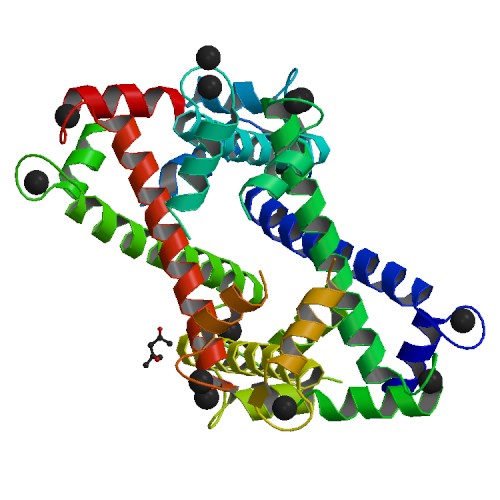 |
Crystal Structure of N-terminal domain EhCaBP1 EF-2 mutant
DOI: 10.2210/pdb5XOP/pdb
Classification: METAL BINDING PROTEIN
Organism(s): Entamoeba histolytica HM-1:IMSS-B
Expression System: Escherichia coli BL21(DE3)
Mutation(s): 5
Deposited: 2017-05-30 Released: 2017-12-06
Deposition Author(s): Kumar, S., Gourinath, S.
Funding Organization(s): BDT BUILDER, UGC-UPE2, DST-PURSE, DST-FIST
|
 |
Crystal Structure of Wild Type Phosphoserine aminotransferase (PSAT) from E. histolytica
DOI: 10.2210/pdb5YB0/pdb
Classification: TRANSFERASE
Organism(s): Entamoeba histolytica
Expression System: Escherichia coli
Deposited: 2017-09-02 Released: 2018-10-31
Deposition Author(s): Singh, R.K., Gourinath, S.
|
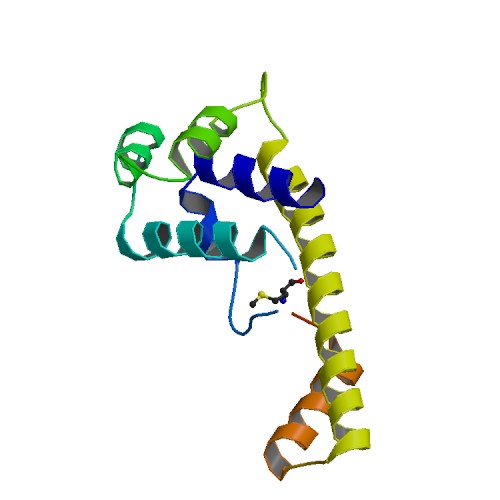 |
Cystal structure of DnaG primase C-terminal domain from Vibrio cholerae
DOI: 10.2210/pdb4IM9/pdb
Classification: TRANSFERASE
Expression System: Escherichia coli
Deposited: 2013-01-02 Released: 2014-05-07
Deposition Author(s): Abdul Rehman, S.A., Tarique, K.F., Gourinath, S.
|
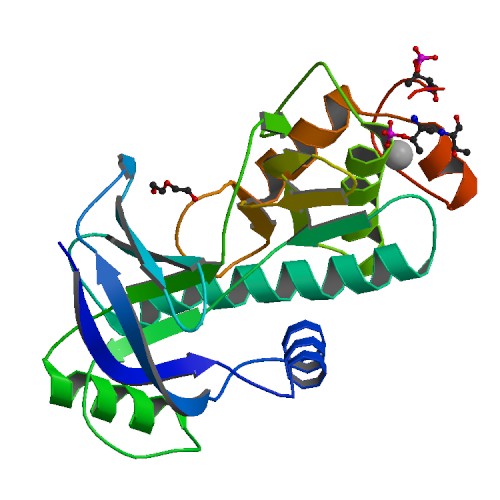 |
Structural and functional characterization of a novel Alpha Kinase from Entamoeba histolytica
DOI: 10.2210/pdb4KUJ/pdb
Classification: TRANSFERASE
Organism(s): Entamoeba histolytica HM-1:IMSS-B
Expression System: Escherichia coli
Deposited: 2013-05-22 Released: 2014-05-28
Deposition Author(s): Rehman, S.A.A., Tarique, K.F., Bhattacharya, A., Gourinath, S.
|
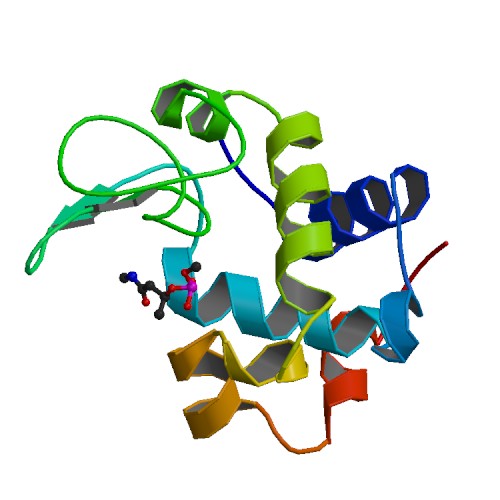 |
Crystal structure of Chicken egg white lysozyme adduct with Organophosphorus pesticide Monochrotophos
DOI: 10.2210/pdb4TUN/pdb
Classification: HYDROLASE
Organism(s): Gallus gallus
Deposited: 2014-06-24 Released: 2014-07-09
Deposition Author(s): Amaraneni, S.R., Kumar, S., Samudrala, G.
|
 |
Crystal structure of Cysteine synthase from Helicobacter pylori
DOI: 10.2210/pdb5HBG/pdbEntry 5HBG supersedes 4R2V
Classification: TRANSFERASE
Organism(s): Helicobacter pylori (strain ATCC 700392 / 26695)
Expression System: Escherichia coli BL21(DE3)
Deposited: 2015-12-31 Released: 2016-01-20
Deposition Author(s): Tarique, K.F., Abdul Rehman, S.A., Gourinath, S.
|
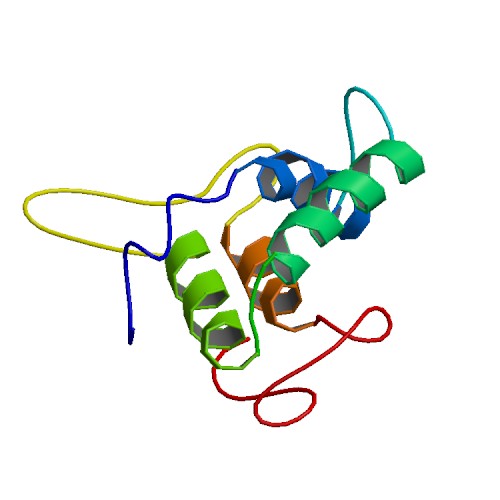 |
Crystal Structure of the Bifunctional Inhibitor Ragi
DOI: 10.2210/pdb1B1U/pdbEntry 1B1U supersedes 1JFO
Classification: HYDROLASE INHIBITOR
Organism(s): Eleusine coracana
Deposited: 1998-11-23 Released: 1998-12-02
Deposition Author(s): Gourinath, S., Srinivasan, A., Singh, T.P.
|
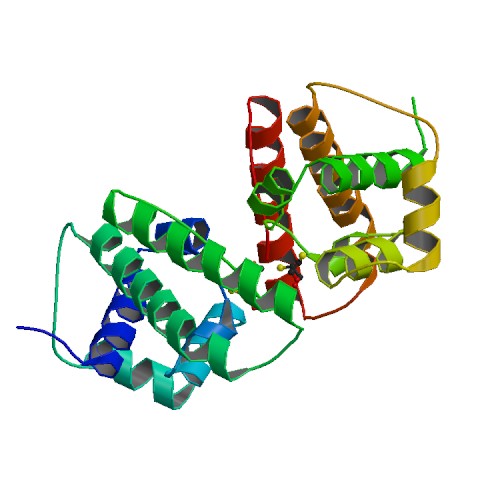 |
Crystal structure of Helicobacter pylori DnaG Primase C terminal domain
DOI: 10.2210/pdb4EHS/pdb
Classification: TRANSFERASE
Organism(s): Helicobacter pylori (strain ATCC 700392 / 26695)
Expression System: Escherichia coli
Deposited: 2012-04-04 Released: 2013-05-01
Deposition Author(s): Abdul Rehman, S.A., Gourinath, S.
|
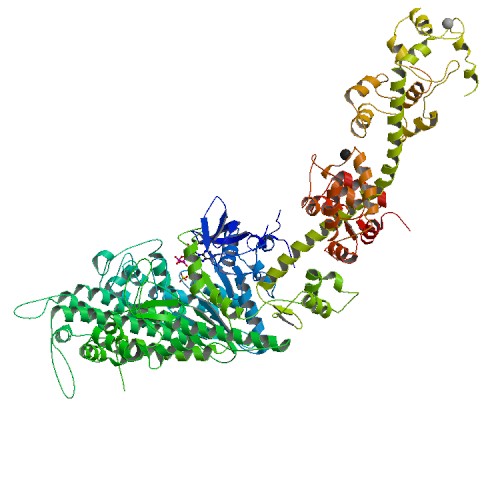 |
Structure of Scallop myosin S1 reveals a novel nucleotide conformation
DOI: 10.2210/pdb1S5G/pdb
Classification: CONTRACTILE PROTEIN
Organism(s): Argopecten irradians
Deposited: 2004-01-20 Released: 2004-06-22
Deposition Author(s): Risal, D., Gourinath, S., Himmel, D.M., Szent-Gyorgyi, A.G., Cohen, C.
|
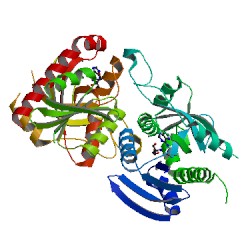 |
Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Bacillus anthracis
DOI: 10.2210/pdb4QEZ/pdb
Classification: HYDROLASE
Organism(s): Bacillus anthracis
Expression System: Escherichia coli
Deposited: 2014-05-19 Released: 2014-06-18
Deposition Author(s): Tarique, K.F., Devi, S., Abdul Rehman, S.A., Gourinath, S.
|
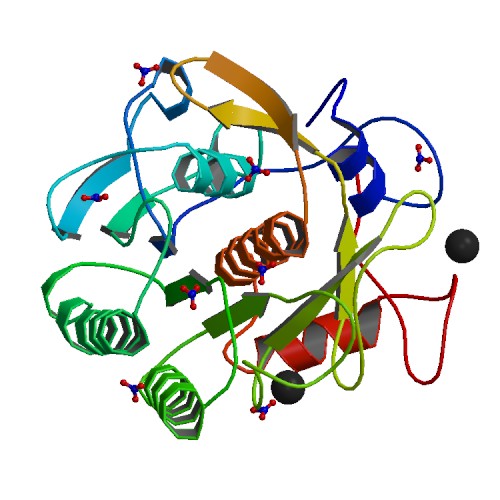 |
Structure of a Serine Protease Proteinase K from Tritirachium Album Limber at 0.98 A Resolution
DOI: 10.2210/pdb1IC6/pdb
Classification: HYDROLASE
Organism(s): Parengyodontium album
Deposited: 2001-03-30 Released: 2001-04-11
Deposition Author(s): Betzel, C., Gourinath, S., Kumar, P., Kaur, P., Perbandt, M., Eschenburg, S., Singh, T.P.
|
×
![]()

